Over the past few years, our group has written several papers on inferring haplotypes from sequence data.
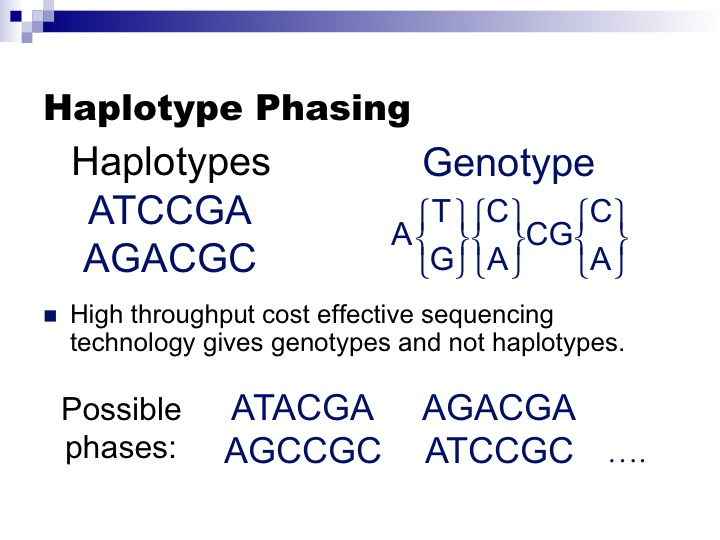
The problem of Haplotype Inference referred to as Haplotype Phasing has had a long history in computational genetics and the problem itself has had several incarnations. Genotyping technologies obtain “genotype” information on SNPs which mixes the genetic information from both chromosomes. However, many genetic analyses require “haplotype” information which is the genetic information on each chromosome (see Figure).
In the early days before reference datasets were available, methods would be applied to large numbers of genotyped individuals which would attempt to identify a small number of haplotypes which explained the majority of the individual genotypes. Methods from this period include PHASE (11254454) and HAP (14988101) (from our group with Eran Halperin). The figure is actually one of Eran’s slides from around 2002.
Once reference datasets such as the HapMap became available, imputation based methods such as IMPUTE(10.1038/ng2088) and BEAGLE(10.1016/j.ajhg.2009.01.005) dominated previous phasing approaches because they leveraged information from the carefully curated reference datasets.
In principal, haplotype phasing or imputation methods can be applied directly to sequencing data by first calling genotypes in the sequencing data and then applying a phasing or imputation approach. However, since each read originates from only one chromosome, if a read spans two genotypes it provides some information on haplotype phase. Combining these reads to construct haplotypes is referred to as the “haplotypes assembly” problem which was pioneered by Vikas Bansal and Vineet Bafna(10.1093/bioinformatics/btn298),(10.1101/gr.077065.108). Dan He in our group developed an optimal method for haplotype assembly which guarantees finding the optimal solution for short reads and reduces the problem of haplotype assembly for longer reads to MaxSAT which finds the optimal solution for the vast majority of problem instances(10.1093/bioinformatics/btq215). More recently, others have developed methods that can discover optimal solutions for all problem instances(10.1093/bioinformatics/btt349). In his paper, Dan also showed that haplotype assembly will always underperform traditional phasing methods for short read sequencing data because too few of the reads span multiple genotypes.
To overcome this issue, Dan extended his methods to jointly perform imputation and haplotype assembly(10.1089/cmb.2012.0091),(10.1016/j.gene.2012.11.093). These methods outperformed both imputation methods and haplotype assembly methods but unfortunately are too slow and memory intensive to apply in practice. More recently, in our group, Wen-Yun Yang, Zhanyong Wang, Farhad Hormozdiari with Bogdan Pasaniuc developed a sampling method which is both fast and accurate for combining haplotype assembly and imputation(10.1093/bioinformatics/btt386).
Full citations of our papers are here: